The Bioinformatics & Genomic Data Quiz!
To round off the Bioinformatics, Statistics, and Data Interpretation in Genomic Analysis module, you can put your new skills to the test - in this challenge quiz, you will take a tour of genomic data resources and extract or compute information to answer the questions.
First - you need to be in a team! Pick your team now, or we can self-organise.
Then to complete the quiz you need this grid:
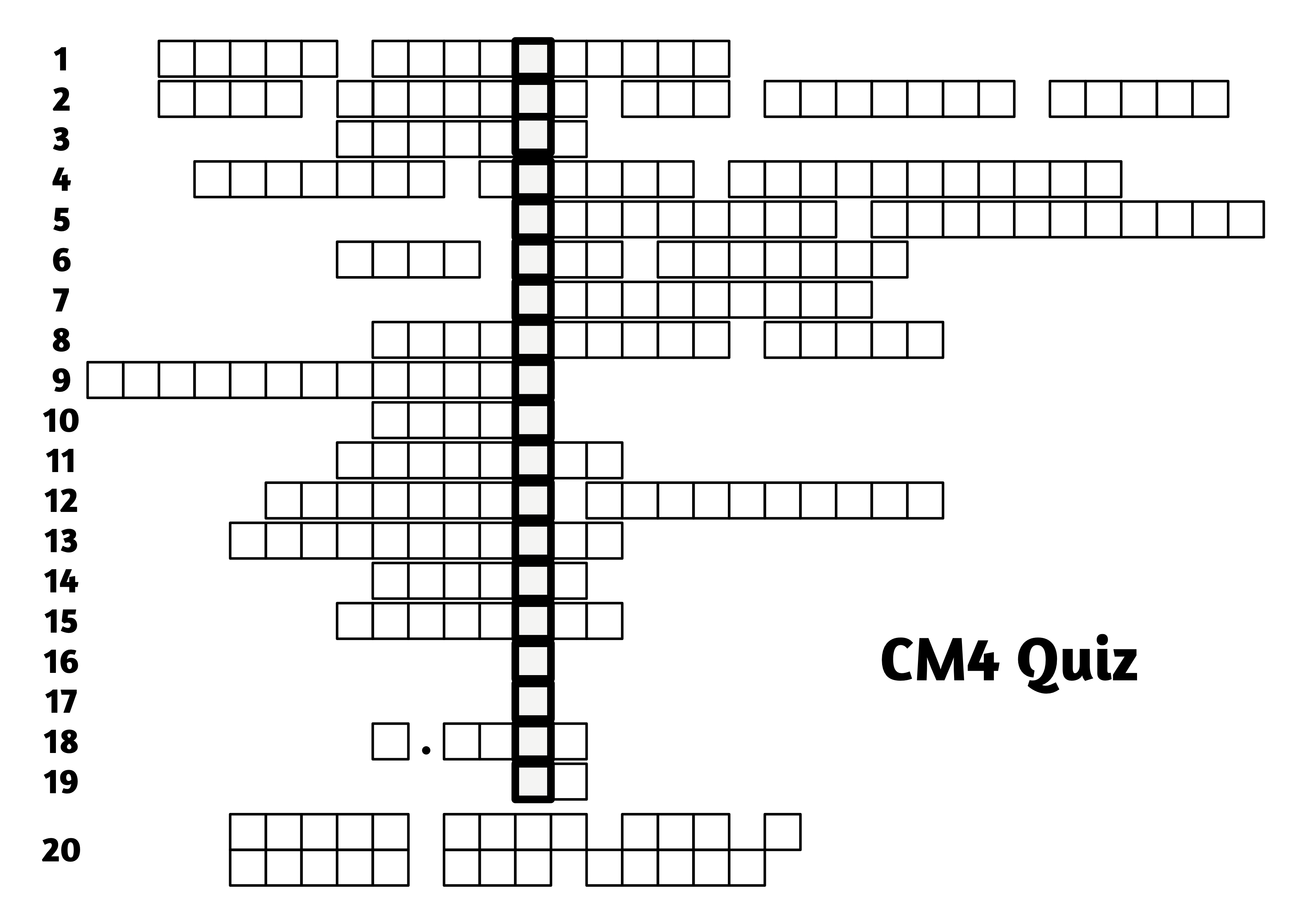
which we've printed for you. Fill in your answers in the grid provided to solve the mystery message.
Rules:
- you can use any tools you like to solve these puzzles.
- first one to complete all the answers and discover the message is the winner!
Good luck!
Clue 1: HBA1
First, we'll delve into the alpha-globin locus on chromosome 16:
Navigate to the UCSC Genome Browser homepage. Let's start by resetting all settings - in the 'Genome Browser' menu at the top, select 'Reset All User Settings'. Now you are ready to start!
Search for 'HBA1' (or 'hba1', it's not case sensitive) in the search box and click the 'go' button to the right
In the list of results, click on the NCBI RefSeq gene identifier for HBA1 - this will take you to the genome browser view focused on the HBA1 gene
Click on the gene in the 'NCBI RefSeq genes' track (either the identifier HBA1/NM_000558.5 or anywhere along the gene) and read the summary information about the gene. (For this question, don't use the GENCODE track which gives slightly different results.)
What does the abbreviation HbF stand for? Enter the answer grid (NB use the American spelling in your answer)
Clue 2: sequence lengths
At the top of the summary page you'll see a link to the RefSeq entry for HBA1 - called NM_000558.5. Click on that now to
visit NCBI RefSeq.
The NCBI Handbook has detailed information about RefSeq annotations in Chapter 18: The Reference Sequence (RefSeq) Database
Let's use RefSeq to get the gene sequence:
Click the FASTA link
Copy and paste the header row and gene sequence and save in a text file (extension can be any of .txt, .fa, .fasta)
Read the content into R.
What is the length of the mRNA sequence for HBA1? Enter this value (in words) into the grid.
- Question
- Hint
The Genomic Data in R tutorial explained how to do this.
Clue 3: getting a specific base
What is the identity of the base at position 280 in the HBA1 mRNA sequence you just loaded? Write the name of the nucleotide base you identify in the grid.
Clue 4: gene identifiers
Look at the table at the link below, which describes various RefSeq identifiers: https://www.ncbi.nlm.nih.gov/books/NBK21091/table/ch18.T.refseq_accession_numbers_and_mole/?report=objectonly
What does the NM_ prefix indicate about the nature of the HBA1 gene?
Write the answer in the grid.
Clue 5: OMIM in the gloamin'
Go back to the genome browser and this time click on the HBA1 gene in the 'GENCODE V44' track rather than the 'RefSeq' track. (Somewhat confusingly, clicking on the gene in different tracks like this gives you slightly different views of the data.)
(If you don't see the 'GENCODE' track - make sure you have clicked 'Reset All User Settings' in the 'Genome Browser' menu at the top of the screen first.)
You should see a page like this:
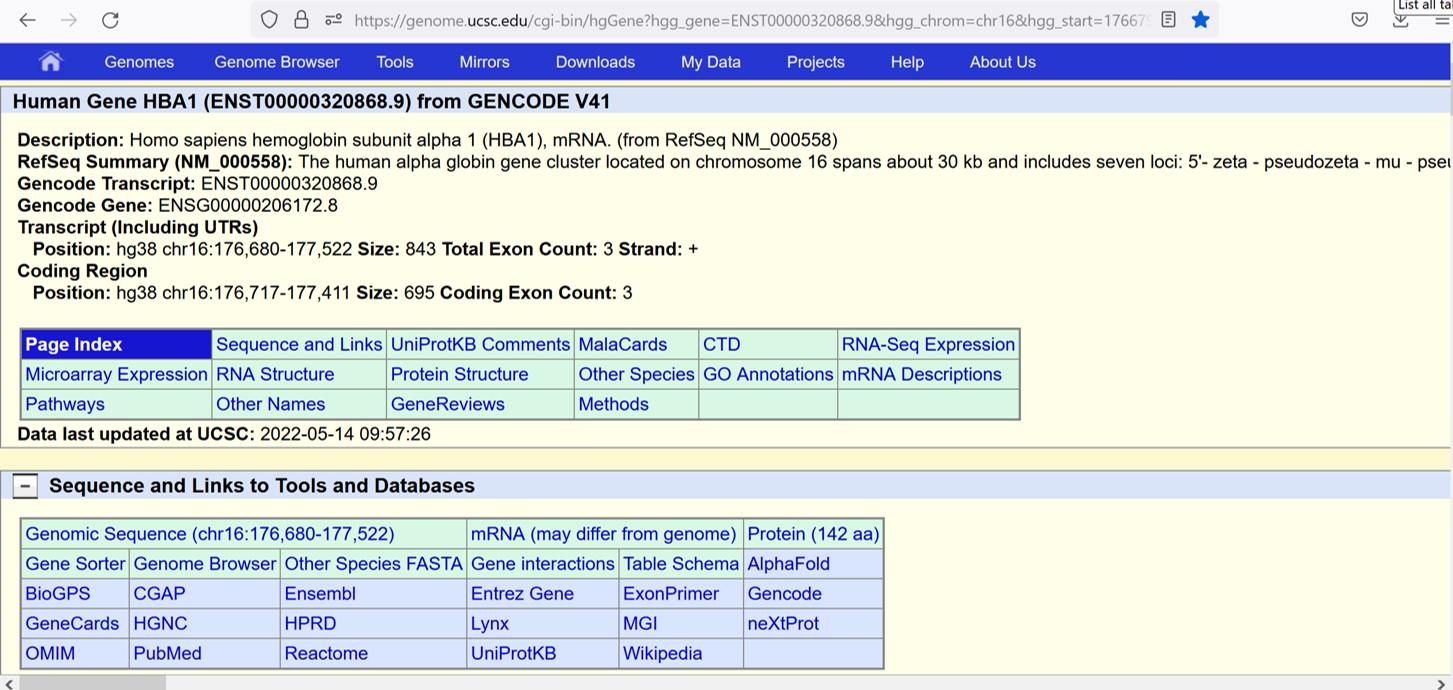
The second table on the page provides links to more than a dozen external tools and databases that provide a rich and comprehensive source of information about the gene, protein, biological pathways and role in disease. Let's look at OMIM:
Click on the OMIM link in the table
Briefly look at the information available for HBA1 to get a sense of the scope and depth of information available
Return to the top of the page and click the green box 'PheneGene Graphics' and select 'radial'
Up pops a very cool graphical representation of HBA1, related genes and their links to disease; the corresponding entry numbers in the OMIM database are shown and directly link to detailed pages by clicking on the gene symbol or disease name. It is also interactive as you can explore related conditions by clicking the OMIM reference number part of the annotations and this will re-draw the map centred on that gene/disease.
What does the acronym OMIM stand for? (It's on this page somewhere!)
Enter the second and third words into your grid.
Clue 6: GOing places
AmiGO 2 is a project to create the next official web-based set of tools for searching and browsing the Gene Ontology database, which is a 'controlled vocabulary' of terms covering biological concepts. A large number of genes or gene products have been annotated using GO terms.
- From the External Links menu on the right-hand side of the OMIM page for HBA1, click 'Gene Info' and follow the 'Gene Ontology' link in the drop-down list.
All GO annotations across different species are listed (132 in total) so let's filter them down a bit:
Scroll down and select 'Organism' on the left; select humans by clicking the green box with a '+' symbol next to 'Homo sapiens'.
Further down, select 'GO class (excluding "regulates")' and 'molecular_function' from the list of filters to add.
There should be 38 annotations in the filtered list. Write down the three-word GO term for the 3rd entry in the results list.
The GO identifier for this term is GO:0005506 (this will appear when the mouse hovers over the GO term). The search can also be done in reverse by entering this ID in the AmiGO homepage to get to the answer another way - most of these databases are updated daily and likely to change over time.
Back in the UCSC genome browser page for HBA1, if you scroll a long way down you will find GO annotation terms for the gene and other things like protein structure information.
Clue 7: on the brain
Recall that HBA1 sits within the human alpha globin gene cluster, which comprises 7 loci.
Go back to your UCSC genome browser tab and click 'back' to return to the browser view (or search again for HBA1) and zoom out until the whole cluster is visible.
Look downstream for the first gene with more than one exon, that is immediately adjacent to the globin cluster (i.e. to the right of HBQ1). To check you have the right gene, its coordinates are chr16:188,969-229,463. (Make sure you are using the
GRCh38/hg38assembly for this quiz.)Inspect the gene expression track just below the transcript depictions for this gene
Click on the bargraph to see more details of how this gene is expressed in different tissues (GTEx track)
This gene has elevated expression in a particular brain region. Which region is it? Write the name in the grid.
Clue 8: where's the gene?
Now let's switch to the Ensembl genome browser and tools to look at a different gene, CFTR. Mutations in CFTR cause cystic fibrosis. Use the Ensembl genome browser to search for CFTR in human genome GRCh38.p13.
What chromosome is CFTR located on? Write down its name in words.
Clue 9: Amino acids
Search online to find out some details about the mutations that cause cystic fibrosis. The most common mutation is a deletion of the 508th amino acid, about halfway along the gene.
Write down the chemical name of the amino acid that is deleted.
You can solve this however you choose - programming or google - but bonus points/kudos if you do it bioinformatically!
Clue 10: The quality of bases
In DNA sequencing, the identification of individual nucleotide bases is assigned a quality score. They are usually represented on a particular scale.
What is the name given to the scale these quality scores are presented on? Write it down in the grid.
Look back at the sequence data analysis tutorial if you are not sure.
The answer is also a homophone for the first name of the Nobel prize-winning scientist who invented the earliest DNA sequencing method...
Clue 11: Covering all the bases
Return to your R session with the HBA1 sequence loaded now.
Use an R function to determine which nucleotide appears most frequently in the HBA1 gene sequence. Write down the name of the resulting nucleotide base in your answer grid.
Clue 12: Disease association
Suppose you conduct a test to see whether a particular genetic variant is causing a disease. You collect a number of cases and controls from hospitals and form a 2x2 table to conduct an association test.
You see a strong signal - an estimated relative risk of four! It looks like a major risk factor.
But now you want to include some covariates to check the result is robust.
What kind of statistical test could you use to do this?
- Question
- Hint 1
The Stats II practical might help.
Clue 13: From association to causality
Thinking about causality - why might you want to include covariates anyway? To deal with possible something factors?
- Question
- Hint 1
The Stats II practical might help here too!.
Clue 14: Team names
The Ensembl gene annotation curation team that shares its name with a city in Cuba will give you the answer to clue 14.
Clue 15: a repeat expansion
Amyotrophic Lateral Sclerosis, also known as Motor Neurone Disease, is a progressive and lethal disease of the nervous system. It has recently been in the news due to the death of Doddie Weir, as well as the death in 2018 of Stephen Hawking, both after a long struggle with the disease.
THe most frequency known cause of ALS is a repeat expansion in the gene C9orf72 as shown in the following diagram from that paper:
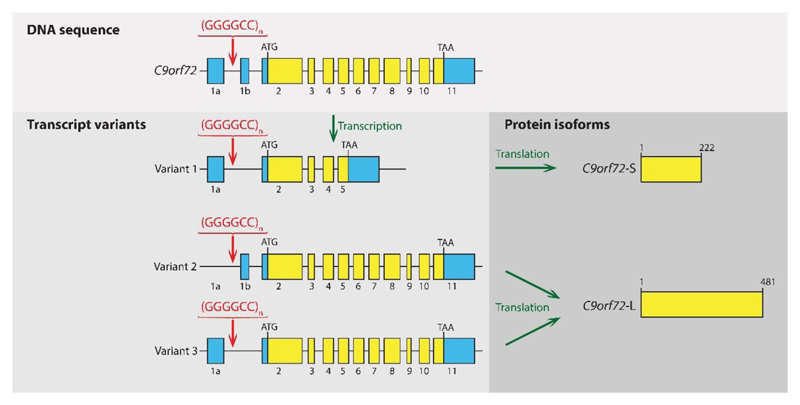
The repeat occurs between the first two exons of (the longest transcript of) C9orf72 - yet despite being noncoding, it is a major risk factor for the disease - as
"...The vast majority (>95%) of neurologically healthy individuals have ≤11 hexanucleotide repeats in the C9orf72 gene [...] an arbitrary cut-off of 30 repeats is used [to determine pathogenicity] in most studies, but larger expansions ranging from hundreds to thousands of repeats are most commonly observed in patients with frontotemporal dementia and ALS."
Use the UCSC genome browser to answer:
- which strand is C9orf72 transcribed on?
- how many copies of the
GGGGCChexanucleotide repeat does the reference sequence carry?
Turning on the 'ClinVar' track under 'Phenotype and literature' may help you find it.
Compute the number of nucleotide bases contained in these repeats. Write that number in words in the grid!
- Question
- Hint 1
- Hint 2
- Hint 3
The repeat is close to the right (upstream) side of the exon at chr9:27,573,426 - 27,573,491.
The 'ClinVar' variants shows clinically-relevant variation - maybe that will help?
The number of nucleotides is (of course) the number of repeat units times the number of bases in the repeat unit
Clue 16 and 17: Counting transcripts
Now let's get back to some command-line programming with gene annotations.
In your (Ubuntu or Mac OS X ) terminal, download or find the file gencode.v41.annotation.gff3.gz from this
folder.
For simplicity gunzip the file first.
Now use the file and your command-line skills (or, if you prefer, your R skills) to answer:
How many transcripts are listed for the gene GYPA ?
Write down the digits in the boxes for clues 16 and 17.
- Question
- Hint 1
- Hint 2
A command like
cat gencode.v41.annotation.gff3 | awk '$3=="transcript"'
will pull out all rows that are transcripts... but how to find those for GYPA?
GYPA encodes glycophorin A, one of the most abundant molecules that gets expressed on the red cell surface. (There are about a million copies on each red cell, and about 25 trillion red cells in the body... that's a lot of glycophorin A!
Clue 18: Counting genes
Using the same file, can you answer:
What proportion of protein-coding genes lie on the X chromosome?
Write your answer (as numbers) to 4 decimal places in the the grid.
- Question
- Hint 1
- Hint 2
- Hint 3 - spoilers!
A command like:
cat gencode.v41.annotation.gff3 | awk '$3=="gene"' | grep 'gene_type=protein_coding'
will find the rows you want. But now you need to count the number of them on each chromosome - how?
Clue 19: Some seasoning
Nearly there! Now download the file 'season.txt' from this folder and explore it using the command line or using R.
How many lines does season.txt contain? Write your answer in the grid.
Clue 20: making edits
You can run this clue in the command-line, in R, or in a text editor - up to you. Working with season.txt, make the following transformations:
- Extract lines 6 and 7 of
season.txt- store them in a new object or file for convenience. - Replace the word "Christmas" with "Xmas"
- Replace "And" with "and"
- concantenate the two lines and print to screen
Enter the last 7 words of the message in your grid.
Congratulations!
You have completed the quiz!
A message should appear in the shaded boxes!