Computational Genomics SRF


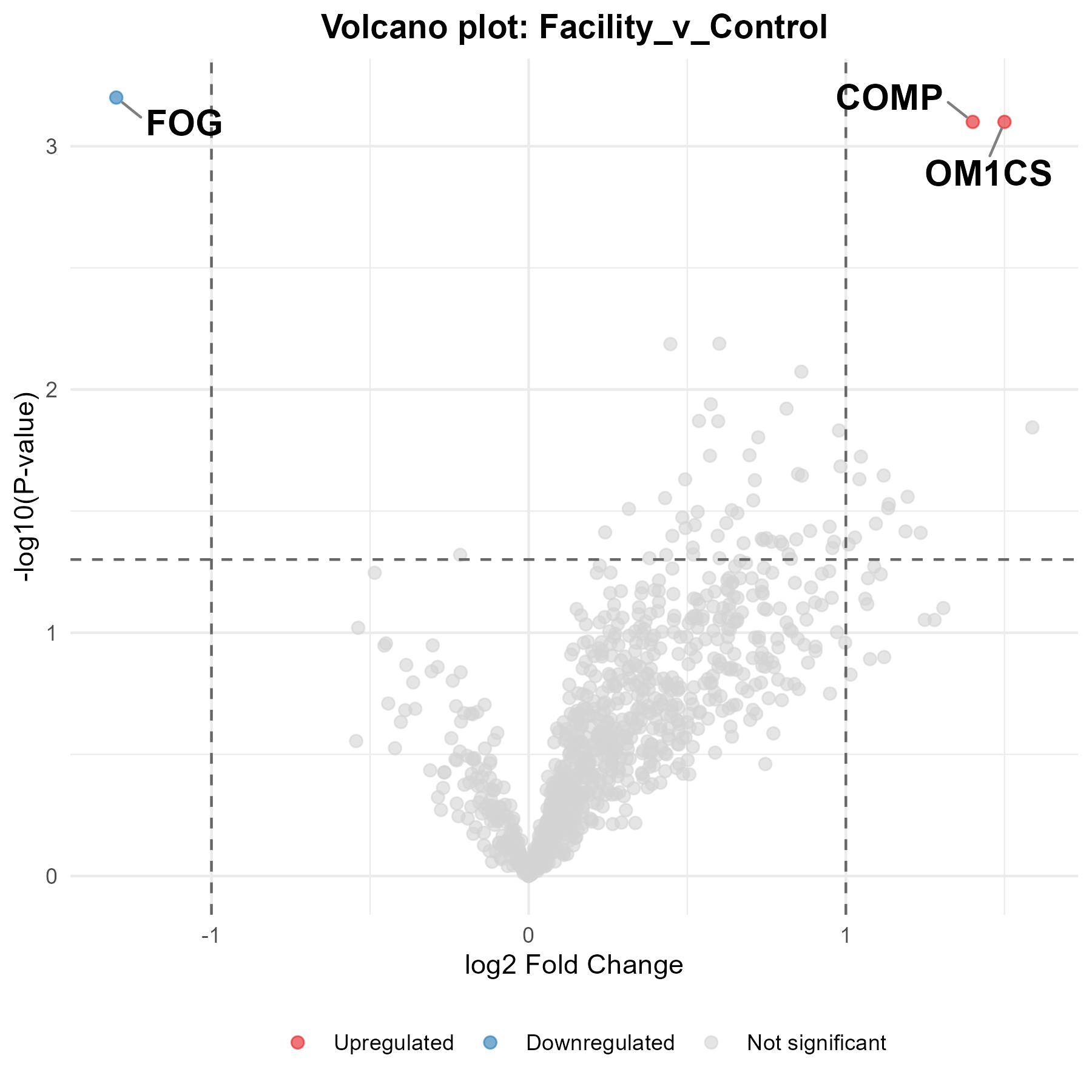
Computational Omics is haunting the halls of this year's Festival of Genomics with the following guest appearances:
Be sure to catch the talk by our sister organisation, Multiomics Platfom Group: High-Resolution Biology: Multi-Dimensional Insights from Spatial Multi-omics (presented by Paolo Piazza) Thursday at 4.45pm
Computational Omics is part of the BioinfoCoreConnect network which will be showcased at the Warwick data science table near the entrance at the Festival of Genomics!
The head of the facility (Matthieu Miossec) will be circulating around the room. Happy to start conversations and collaborations!
The Facility
Deriving meaningful insights from the gigabytes of data coming off high-throughput omics platforms requires a suite of computational tools for transforming, annotating, quantifying and visualising said data. Such detailed analyses require the intervention of dedicated computational biologists with the computing, statistical and biology expertise to turn raw data into biological discoveries.
Located within the Centre for Human Genetics, an integral part of Oxford University's Nuffield Department of Medicine, the Computational Genomics SRF provides standardised and cutting-edge data analysis solutions for a whole host of omics platforms (see below) as well as training and support for those researchers; staff and student, eager to attempt these analyses themselves.
This service is available to all Oxford University research groups and is available to external collaborators at well for an additional cost.
For prices and all other enquiries: compgen@ndm.ox.ac.uk
We offer two services, priced per half day of dedicated computational biologist work:
- support and training (you do the analysis, we assist you with various aspects)
- standard analysis (we do the analysis for you, you get results!)
The following analyses are available at fixed rates based on the standard analysis service rates:
- Bulk RNAseq analysis (from QC and mapping to differential expression, with final report)
- Single-cell RNAseq analysis (from feature-barcode counts to variable feature identification and clustering, with final report)
Note that these rates do not vary with sample number as computational biologist time is not affected by cohort size. Large studies are eagerly welcome!
Some of the omics technologies we have previously worked with are as follows:
- Genomics
- Whole exome
- Whole genome
- Transcriptomics
- Bulk RNAseq
- Single-cell RNAseq
- 10X Visium and Xenium (spatial) [also some Merscope]
- Other omics
- ATAC-seq, CHiP-seq, Olink and Alamar (proteomics)...
though our service is not limited to these technologies, so please do contact us: compgen@ndm.ox.ac.uk
Note: We do not charge for initial meetings or discussions relating to grant application, please don't hesitate to contact us so we can get this discussion going.
Further note: As outlined below, co-authorship is expected as acknowledgment for work performed by our group and we will therefore not accept offers to collaborate for free in exchange for authorship. We will not be able to fulfil any requests without prior payment.
Policies and guidelines FOR publications
As per commonly-accepted scientific practice, we ask that the relevant member(s) of CGG be listed as co-authors in all publications arising from the data.
In such cases, the CGG analyst(s) will provide input for writing and editing co-authored manuscripts, as well as support for the upload of the datasets on a publicly available database repository of high-throughput data, where needed.

